Please fill out the following fields: job name, email, sequence
One or more parameter you entered is not within range, gRNA length should be between 4 and 50, homology arm length should be between 50 and 500, identity should be between 0 and 1 coverage should be between 0 to 1
PAM length should be between 3 and 10, and containing only letters "A", "T", "C", "G", "N", no space(s).
To search gRNa off-targets in your custom genome, please upload Fasta format genome sequence file, 200MB size limit. Please contact us if you wish to add a frequently used genome to our web-server
please paste your input sequence to search gRNA in. Plain sequence or FASTA format.
plain sequence example:
ATGAGCGTAGCTGTGTGCATGTCAGTCGATGGGCATGCATGCATGCAT
Fasta formate example:
>Tc00.1047053503403.20-trans-sialidase
ATGCTCTCACGTGTTGATGCTGTCAAGGCACCCCGCACACACAACCGTCGCCGCGTGACCGGATCCAGCGGAAGGAGGAGGGAAGGAAGAGAGAGTGAGCCGCAGAGGCCCAACATGTCCCGGCATCACTTCTATTCTGCGGTGCTGCTCCTCCTCGTCGTGATGATGTGCCGCGGCAGTGGAGCTGCGCACGCTTTGGAGAGAAACTCAGGGGATTTGCAAATGCCGCAGGAGATCGCCATGTTTGTGCCGA
Note that sequence name will be truncated at space or special characters: ~!?@#$%^&*(){}[]"'/\,;:
An example consisting of 3 fasta-format sequences have been loaded, please keep T. cruzi genome selected to run the example
A multiple copy gene example has been loaded, this gene has >65 copies in the T. cruzi genome, please keep T. cruzi genome selected to run the example
"load example" will load 3 fasta-format sequences fron the T. cruzi CL strain genome
When determining gRNA on-targets, our program automatically compares the homology of each genomic hit to your gRNA-containing sequence. Sufficient homology would render a genomic hit to be an on-target. The homology arm length determines the length of the regions being compared. For example, homology arm length=50, 50bp upstream of gRNA and 50bp downstream of gRNA, plus gRNA (default 23bp), a total of 123bp will be used to determine homology
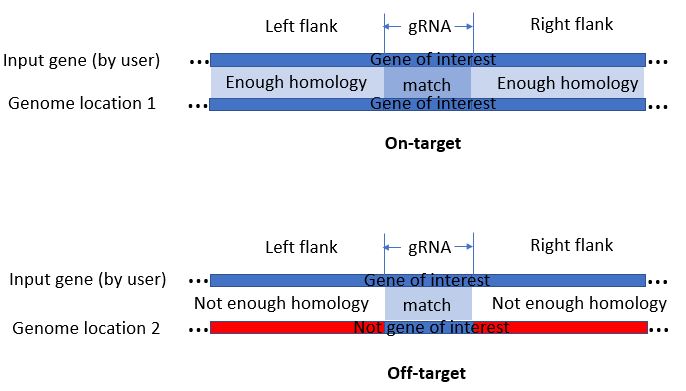
When determining guide RNA on-targets, our program automatically compares the homology of each genomic hit to your guide RNA-containing sequence. Sufficient homology would render a genomic hit to be an on-target. In such process, the identity parameter is the threshold for minimum alignment identity between genomic hits and guide RNA-containing sequence.

When determining guide RNA on-targets, our program automatically compares the homology of each genomic hit to your guide RNA-containing sequence. Sufficient homology would render a genomic hit to be an on-target. In such process, the coverage parameter is the threshold for minimum alignment coverage between genomic hits and guide RNA-containing sequence.

You selected a custom genome file to upload, but you didn't select the custom genome option, please select the "upload your custom genome" option under "custom genome" section and click "Get gRNA" again.
You selected the option to upload a custom genome file, but you didn't select anyfile.
For any gene sequences that is less than 100bp, EuPaGDT won't be able to accurately assess gRNA on-targets in the genome .
When determining gRNA off-targets, a genomic sequence will be considered potential off-target if the alignment of it with gRNA has less than n mismatches. "n" is the "maximum number of mismatch" parameter value
RNA-guided nucleases such as Cas9 can cleave targets using alternative PAMs at lower efficiency.
For example SpCas9 can also use PAM "NAG", "NGA"(with lower efficiency compared to "NGG");
gRNA "ATCGATCGATCGATCGATCG|NGG" could have the off-targets:
"ATCGATCGATCGATCGATCG|NAG",
"ATCGATCGATCGATCGATCG|NGA"
in undesired loci.
This is the number of top-ranking gRNAs will be chosen for each of your input sequence.
Please do not include special characters such as !@#$%^&().,; in job names
parameter for "% of length of input sequence" should be between 1 and 100
In some scenarios such as gene knockout experiment, it is preferred to disrupt genes at positions closer to start codon to prevent production of truncated protein that may have partial functionality.
The following parameters will be automatically set if respective nuclease is chosen:
SpCas9: gRNA length 20; 3'PAM: NGG; off-target PAM: NAG, NGA
SaCas9: gRNA length 21; 3'PAM: NNGRRT; off-target PAM: NNGRRA,NNGRRG,NNGRRC
AsCpf1: gRNA length 20; 5'PAM: TTTN; off-target PAM: None
CjCas9: gRNA length 23; 3'PAM: NNNNACA; off-target PAM: NNNNACAC,NNNNRYAC
FnCas9: gRNA length 21; 3'PAM: NGG; off-target PAM: NGA,NAG,NAA
FnCpf1: gRNA length 23; 5'PAM: TTN,YG; off-target PAM: CTN
LbCpf1: gRNA length 20; 5'PAM: TTTN; off-target PAM: None
NmCas9: gRNA length 20; 3'PAM: NNNNGANN; off-target PAM: NNNNGTTN,NNNNGNNT,NNNNGTNN,NNNNGNTN
St1Cas9: gRNA length 20; 3'PAM: NNAGAA,NNGGAA; off-target PAM: NNAGGA,NNANAA,NNGGGA
TdCas9: gRNA length 20; 3'PAM: NAAAAC; off-target PAM: None
Conserved regions are identified by tblastx (translated query vs translated database) input sequence against the following 13 genomes:
Mus Musculus,
S.cerevisiae S288c,
D. melanogaster r6.10,
N. crassa OR74A,
E. histolytica HM1IMSS-B,
C. parvum IowaII,
P. falciparum 3D7,
T. gondii ME49,
T. cruzi CLBrener,
L. donovani BPK282A1,
G. intestinalis AssemblageA,
E. cuniculi GBM1,
T. annulata Ankara.
A region of input sequence is marked as conserved region if tblastx-matches are found for at least 12 out of 13 genomes.
Please use IUPAC code for degenerative base(s) in PAM motif
| IUPAC nucleotide code | Base |
| A | Adenine |
| C | Cytosine |
| G | Guanine |
| T | Thymine |
| R | A or G |
| Y | C or T |
| S | G or C |
| W | A or T |
| K | G or T |
| M | A or C |
| B | C or G or T |
| D | A or G or T |
| H | A or C or T |
| V | A or C or G |
| N | any base |
Reference:
IUPAC-IUB SYMBOLS FOR NUCLEOTIDE NOMENCLATURE:
Cornish-Bowden (1985) Nucl. Acids Res. 13: 3021-3030.
Eukaryotic Pathogen CRISPR guide RNA/DNA Design Tool
with (1) custom genome upload, (2) off-target analysis, (3) on-targets searching (for targeting gene families), (4) efficiency/activity prediction, (5) assisted oligo repair template design , (6) gRNA transcription problem identification, (7) flanking microhomology searching (for predicting deletions)
** Batch Mode **
upload and process multiple sequences in one run
if you are processing one gene, please use this link
A multi-thread version of batch mode is available upon request for analyzing >300 sequences
(please use Chrome browser or enable Javascript in your browser for optimal results)



